人类的发展史就是和各种疾病争斗的发展史。新冠肺炎对我们的工作和生活产生巨大的影响,通过数据分析,来了解下疫情的发展情况如何。数据来源于这个github仓库。
1 读取数据
library(tidyplots)
dinosaurs %>%
# 依次添加条形图、线、点、值和连线
tidyplot(x = time_lived) %>%
add_count_bar(alpha = 0.4) %>%
add_count_dash() %>%
add_count_dot() %>%
add_count_value() %>%
add_count_line() %>%
# 旋转x轴标签
adjust_x_axis(rotate_labels = TRUE) %>%
# 调整图形颜色
adjust_colors(c("black", "white")) %>%
theme_ggplot2() %>%
add(ggplot2::theme(axis.text.x = ggplot2::element_text(
size = 4, color = "red", angle = 45, hjust = 1, vjust = 1
))) %>%
# 移除x轴的刻度线
remove_x_axis_ticks() %>%
# 移除x轴的标题
remove_x_axis_title()
df <- read_csv("D:/Myblog/datas/time_series_covid19_confirmed_global.csv")
df %>%
slice_head(n = 6)# A tibble: 6 × 74
`Province/State` `Country/Region` Lat Long `1/22/20` `1/23/20` `1/24/20`
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 <NA> Afghanistan 33 65 0 0 0
2 <NA> Albania 41.2 20.2 0 0 0
3 <NA> Algeria 28.0 1.66 0 0 0
4 <NA> Andorra 42.5 1.52 0 0 0
5 <NA> Angola -11.2 17.9 0 0 0
6 <NA> Antigua and Barbu… 17.1 -61.8 0 0 0
# ℹ 67 more variables: `1/25/20` <dbl>, `1/26/20` <dbl>, `1/27/20` <dbl>,
# `1/28/20` <dbl>, `1/29/20` <dbl>, `1/30/20` <dbl>, `1/31/20` <dbl>,
# `2/1/20` <dbl>, `2/2/20` <dbl>, `2/3/20` <dbl>, `2/4/20` <dbl>,
# `2/5/20` <dbl>, `2/6/20` <dbl>, `2/7/20` <dbl>, `2/8/20` <dbl>,
# `2/9/20` <dbl>, `2/10/20` <dbl>, `2/11/20` <dbl>, `2/12/20` <dbl>,
# `2/13/20` <dbl>, `2/14/20` <dbl>, `2/15/20` <dbl>, `2/16/20` <dbl>,
# `2/17/20` <dbl>, `2/18/20` <dbl>, `2/19/20` <dbl>, `2/20/20` <dbl>, …# 查看数据结构
glimpse(df)Rows: 256
Columns: 74
$ `Province/State` <chr> NA, NA, NA, NA, NA, NA, NA, NA, "Australian Capital T…
$ `Country/Region` <chr> "Afghanistan", "Albania", "Algeria", "Andorra", "Ango…
$ Lat <dbl> 33.0000, 41.1533, 28.0339, 42.5063, -11.2027, 17.0608…
$ Long <dbl> 65.0000, 20.1683, 1.6596, 1.5218, 17.8739, -61.7964, …
$ `1/22/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ `1/23/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ `1/24/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ `1/25/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ `1/26/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 3, 0, 0, 0, 0, 1, 0, 0, 0,…
$ `1/27/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 0, 0, 0, 1, 0, 0, 0,…
$ `1/28/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 0, 0, 0, 1, 0, 0, 0,…
$ `1/29/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 1, 0, 0, 1, 0, 0, 0,…
$ `1/30/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 3, 0, 0, 2, 0, 0, 0,…
$ `1/31/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 2, 0, 0, 3, 0, 0, 0,…
$ `2/1/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 3, 1, 0, 4, 0, 0, 0,…
$ `2/2/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 2, 2, 0, 4, 0, 0, 0,…
$ `2/3/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 2, 2, 0, 4, 0, 0, 0,…
$ `2/4/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 3, 2, 0, 4, 0, 0, 0,…
$ `2/5/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 3, 2, 0, 4, 0, 0, 0,…
$ `2/6/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 4, 2, 0, 4, 0, 0, 0,…
$ `2/7/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/8/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/9/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/10/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/11/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/12/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/13/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/14/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/15/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/16/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/17/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/18/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/19/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/20/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/21/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/22/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/23/20` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/24/20` <dbl> 1, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 0, 0,…
$ `2/25/20` <dbl> 1, 0, 1, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 2, 0,…
$ `2/26/20` <dbl> 1, 0, 1, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 2, 0,…
$ `2/27/20` <dbl> 1, 0, 1, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 3, 0,…
$ `2/28/20` <dbl> 1, 0, 1, 0, 0, 0, 0, 0, 0, 4, 0, 5, 2, 0, 4, 0, 3, 0,…
$ `2/29/20` <dbl> 1, 0, 1, 0, 0, 0, 0, 0, 0, 4, 0, 9, 3, 0, 7, 2, 9, 0,…
$ `3/1/20` <dbl> 1, 0, 1, 0, 0, 0, 0, 1, 0, 6, 0, 9, 3, 0, 7, 2, 14, 3…
$ `3/2/20` <dbl> 1, 0, 3, 1, 0, 0, 0, 1, 0, 6, 0, 9, 3, 1, 9, 2, 18, 3…
$ `3/3/20` <dbl> 1, 0, 5, 1, 0, 0, 1, 1, 0, 13, 0, 11, 3, 1, 9, 2, 21,…
$ `3/4/20` <dbl> 1, 0, 12, 1, 0, 0, 1, 1, 0, 22, 1, 11, 5, 1, 10, 2, 2…
$ `3/5/20` <dbl> 1, 0, 12, 1, 0, 0, 1, 1, 0, 22, 1, 13, 5, 1, 10, 3, 4…
$ `3/6/20` <dbl> 1, 0, 17, 1, 0, 0, 2, 1, 0, 26, 0, 13, 7, 1, 10, 3, 5…
$ `3/7/20` <dbl> 1, 0, 17, 1, 0, 0, 8, 1, 0, 28, 0, 13, 7, 1, 11, 3, 7…
$ `3/8/20` <dbl> 4, 0, 19, 1, 0, 0, 12, 1, 0, 38, 0, 15, 7, 2, 11, 3, …
$ `3/9/20` <dbl> 4, 2, 20, 1, 0, 0, 12, 1, 0, 48, 0, 15, 7, 2, 15, 4, …
$ `3/10/20` <dbl> 5, 10, 20, 1, 0, 0, 17, 1, 0, 55, 1, 18, 7, 2, 18, 6,…
$ `3/11/20` <dbl> 7, 12, 20, 1, 0, 0, 19, 1, 0, 65, 1, 20, 9, 3, 21, 9,…
$ `3/12/20` <dbl> 7, 23, 24, 1, 0, 0, 19, 4, 0, 65, 1, 20, 9, 3, 21, 9,…
$ `3/13/20` <dbl> 7, 33, 26, 1, 0, 1, 31, 8, 1, 92, 1, 35, 16, 5, 36, 1…
$ `3/14/20` <dbl> 11, 38, 37, 1, 0, 1, 34, 18, 1, 112, 1, 46, 19, 5, 49…
$ `3/15/20` <dbl> 16, 42, 48, 1, 0, 1, 45, 26, 1, 134, 1, 61, 20, 6, 57…
$ `3/16/20` <dbl> 21, 51, 54, 2, 0, 1, 56, 52, 2, 171, 1, 68, 29, 7, 71…
$ `3/17/20` <dbl> 22, 55, 60, 39, 0, 1, 68, 78, 2, 210, 1, 78, 29, 7, 9…
$ `3/18/20` <dbl> 22, 59, 74, 39, 0, 1, 79, 84, 3, 267, 1, 94, 37, 10, …
$ `3/19/20` <dbl> 22, 64, 87, 53, 0, 1, 97, 115, 4, 307, 1, 144, 42, 10…
$ `3/20/20` <dbl> 24, 70, 90, 75, 1, 1, 128, 136, 6, 353, 3, 184, 50, 1…
$ `3/21/20` <dbl> 24, 76, 139, 88, 2, 1, 158, 160, 9, 436, 3, 221, 67, …
$ `3/22/20` <dbl> 40, 89, 201, 113, 2, 1, 266, 194, 19, 669, 5, 259, 10…
$ `3/23/20` <dbl> 40, 104, 230, 133, 3, 3, 301, 235, 32, 669, 5, 319, 1…
$ `3/24/20` <dbl> 74, 123, 264, 164, 3, 3, 387, 249, 39, 818, 6, 397, 1…
$ `3/25/20` <dbl> 84, 146, 302, 188, 3, 3, 387, 265, 39, 1029, 6, 443, …
$ `3/26/20` <dbl> 94, 174, 367, 224, 4, 7, 502, 290, 53, 1219, 12, 493,…
$ `3/27/20` <dbl> 110, 186, 409, 267, 4, 7, 589, 329, 62, 1405, 12, 555…
$ `3/28/20` <dbl> 110, 197, 454, 308, 5, 7, 690, 407, 71, 1617, 15, 625…
$ `3/29/20` <dbl> 120, 212, 511, 334, 7, 7, 745, 424, 77, 1791, 15, 656…
$ `3/30/20` <dbl> 170, 223, 584, 370, 7, 7, 820, 482, 78, 2032, 15, 689…
$ `3/31/20` <dbl> 174, 243, 716, 376, 7, 7, 1054, 532, 80, 2032, 17, 74…观察发现,数据集中存在缺失值、列名不好理解等问题。
2 数据清洗
- 除1-4列外,其余列名都是日期。
- 我们希望将数据清洗成两列,第一列为日期,第二列为该日期的确诊病例数。
根据此思路,我们对数据进行整理。
df1 <- df %>%
pivot_longer(
cols = 5:ncol(.),
names_to = "date",
values_to = "cases"
) %>%
mutate(date = lubridate::mdy(date)) %>%
janitor::clean_names() %>%
group_by(country_region, date) %>%
summarise(cases = sum(cases)) %>%
ungroup()
df1# A tibble: 12,600 × 3
country_region date cases
<chr> <date> <dbl>
1 Afghanistan 2020-01-22 0
2 Afghanistan 2020-01-23 0
3 Afghanistan 2020-01-24 0
4 Afghanistan 2020-01-25 0
5 Afghanistan 2020-01-26 0
6 Afghanistan 2020-01-27 0
7 Afghanistan 2020-01-28 0
8 Afghanistan 2020-01-29 0
9 Afghanistan 2020-01-30 0
10 Afghanistan 2020-01-31 0
# ℹ 12,590 more rows计算每日确诊病例
df1 %>%
group_by(date) %>%
summarise(confirmed = sum(cases)) %>%
ggplot(aes(x = date, y = confirmed)) +
geom_point() +
scale_x_date(
date_labels = "%m-%d",
date_breaks = "week"
) +
scale_y_continuous(
breaks = c(0, 50000, 100000, 200000, 300000, 500000, 900000),
labels = scales::comma # 不使用科学计数法
) +
theme_bw()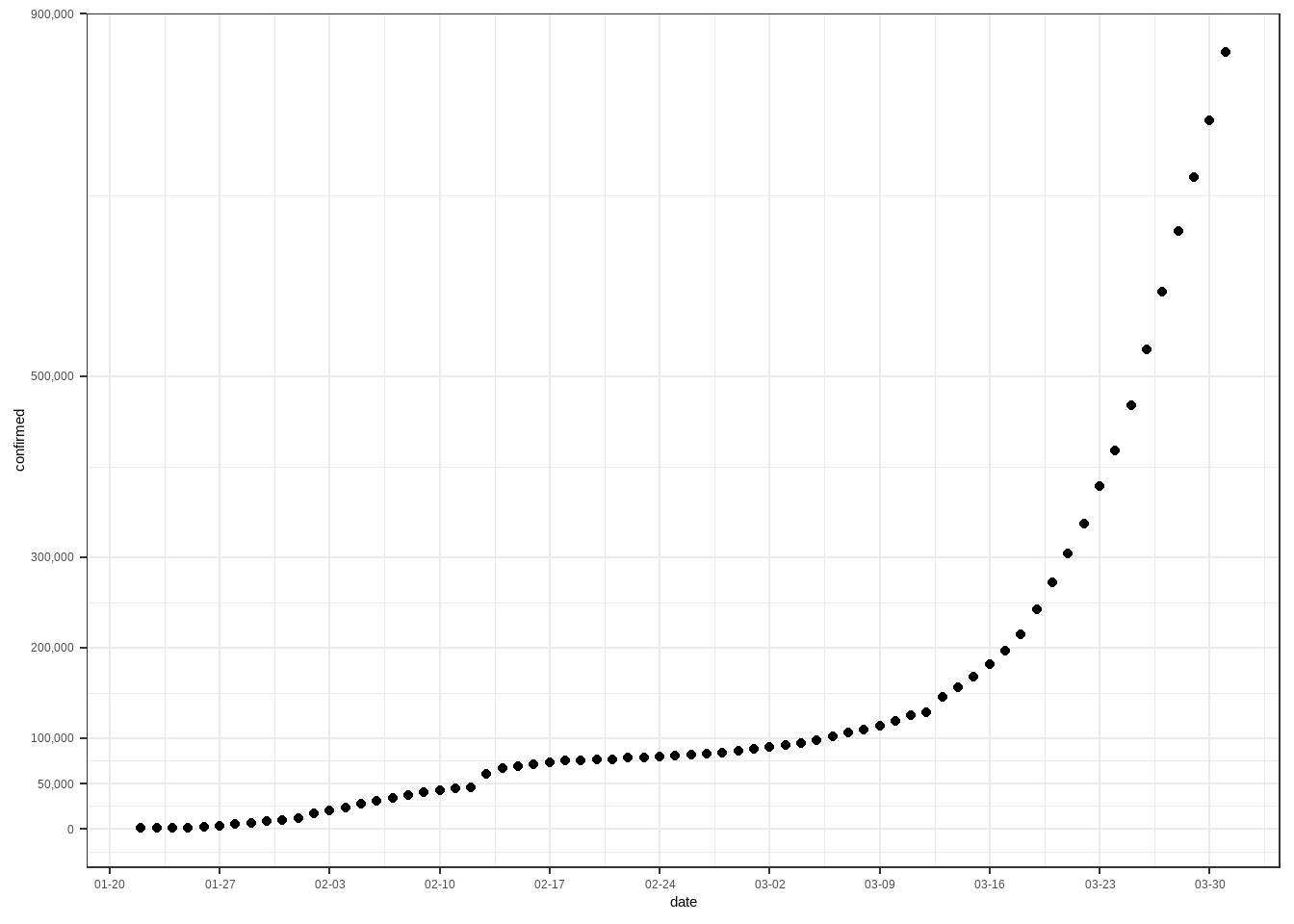
看图说话,从2020年1月20日至3月底,新冠肺炎确诊病例呈快速增长的趋势,而且越来越快。看看咱们国家的情况。
df1 %>%
filter(country_region == "China") %>%
ggplot(aes(x = date, y = cases)) +
geom_point() +
scale_x_date(
date_breaks = "week",
date_labels = "%m-%d"
)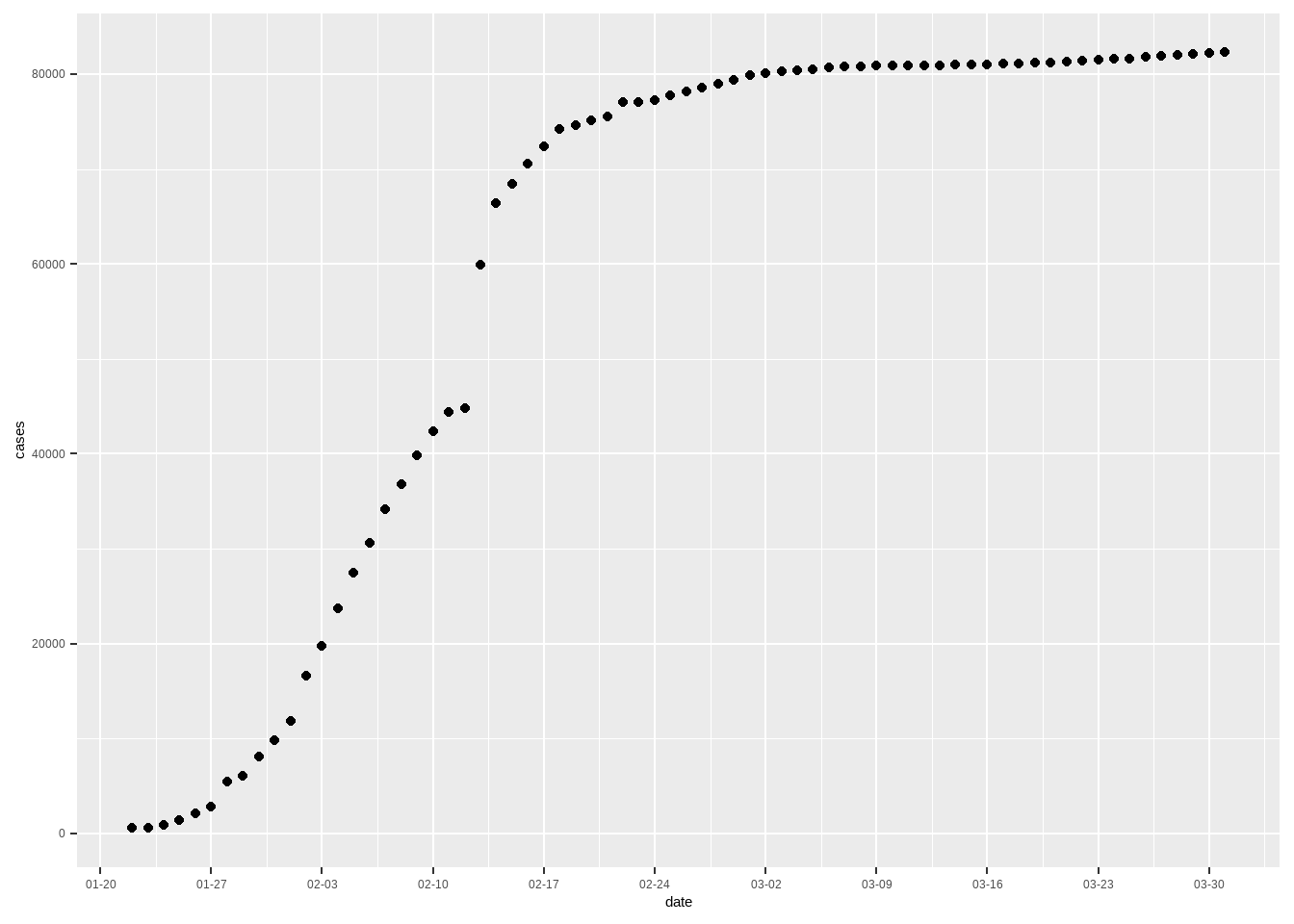
我国在经历短暂病例数增加的后,疫情马上稳定了下来。
df1 %>%
group_by(country_region) %>%
filter(max(cases) >= 20000) %>%
ungroup() %>%
ggplot(aes(x = date, y = cases, color = country_region)) +
geom_point() +
scale_x_date(
date_labels = "%m-%d",
date_breaks = "week"
) +
scale_y_log10() +
facet_wrap("country_region", ncol = 2) +
theme(
axis.text.x = element_text(angle = 45, hjust = 1),
legend.position = "none"
)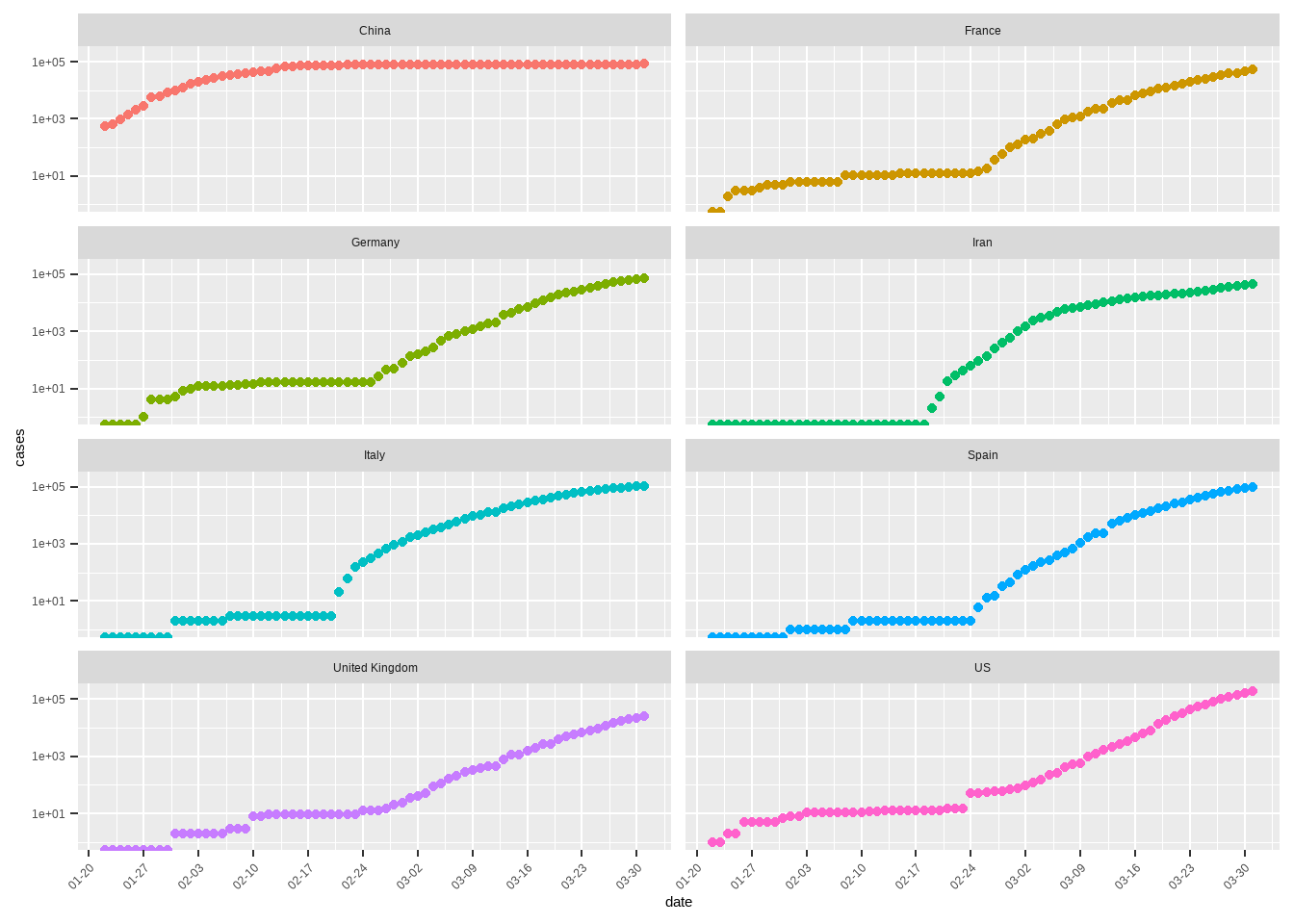
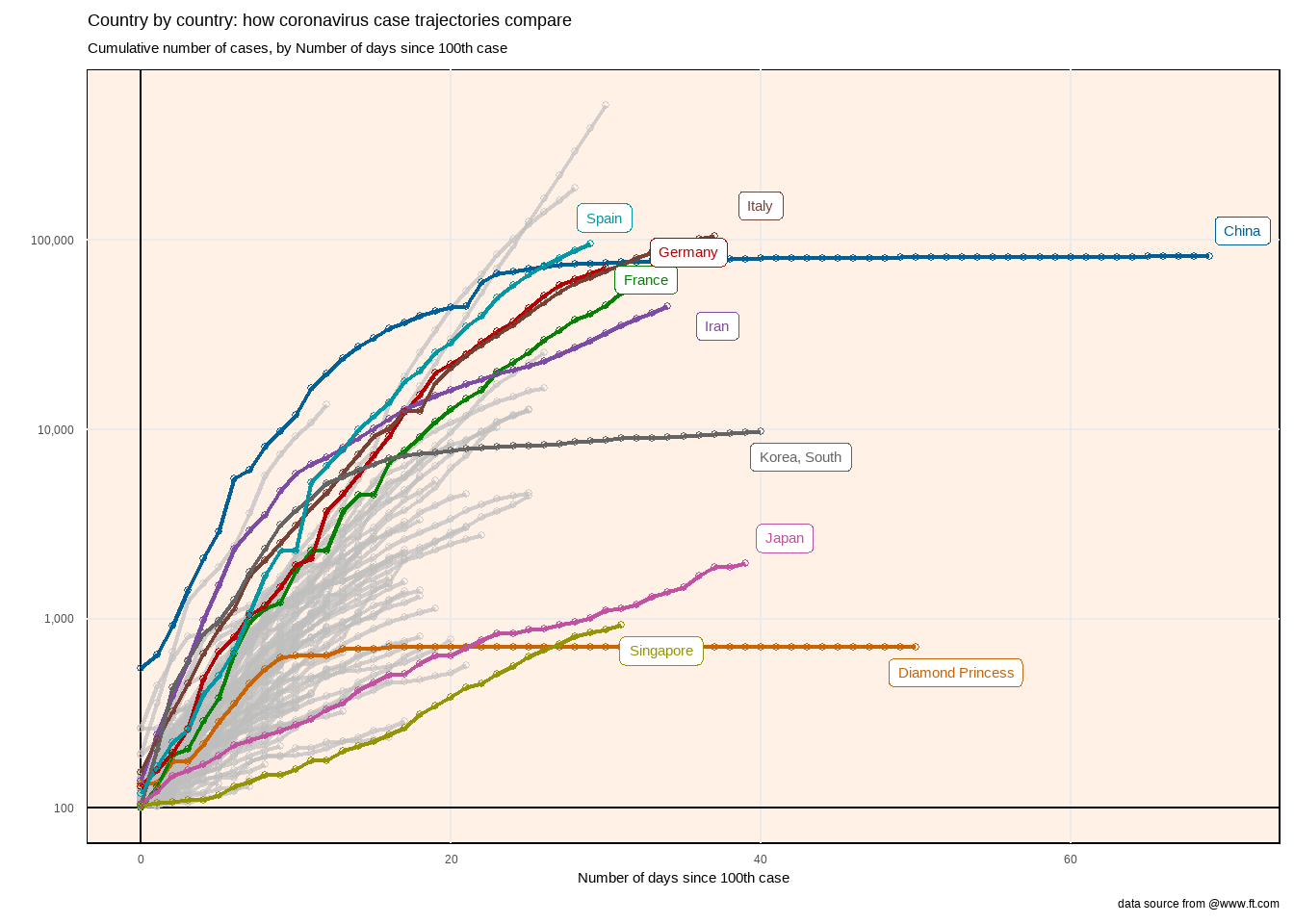
在最大病例数超过20000例的国家中,除我国外,其他国家的病例数都经历了一个断崖式的增长。
3 可视化探索
想要在图形中显示出现100个病例后,各国确认人数的爆发趋势:
- 横坐标为日期,即在出现100个病例后的第几天。
- 纵坐标是累积确诊人数。
要达到这个这个效果,我们需要针对时间轴做一些变形:
- 按国家分组
- 筛选累积确诊数超过100的国家
- 找到所有case超过100的日期,并找到最早达到100病例数的日期(第0天),将其设置为0
- 构建新列,为当前日期与第0天的差值,即从100个病例开始至今过了多少天,并将这列转变为数值。
df2 <- df1 %>%
group_by(country_region) %>%
filter(max(cases) >= 100) %>%
mutate(
days_since_100 = date - min(date[cases >= 100])
) %>%
mutate(days_since_100 = as.numeric(days_since_100)) %>%
filter(days_since_100 >= 0) %>%
ungroup()
df2# A tibble: 1,710 × 4
country_region date cases days_since_100
<chr> <date> <dbl> <dbl>
1 Afghanistan 2020-03-27 110 0
2 Afghanistan 2020-03-28 110 1
3 Afghanistan 2020-03-29 120 2
4 Afghanistan 2020-03-30 170 3
5 Afghanistan 2020-03-31 174 4
6 Albania 2020-03-23 104 0
7 Albania 2020-03-24 123 1
8 Albania 2020-03-25 146 2
9 Albania 2020-03-26 174 3
10 Albania 2020-03-27 186 4
# ℹ 1,700 more rowsdf2_most <- df2 %>%
group_by(country_region) %>%
slice_max(days_since_100, n = 1) %>%
filter(cases >= 10000) %>%
ungroup() %>%
arrange(desc(cases))
highlight <- df2 %>%
group_by(country_region) %>%
slice_max(days_since_100, n = 1) %>%
ungroup() %>%
arrange(desc(cases)) %>%
slice_max(days_since_100, n = 10) %>%
pull(country_region)df2 %>%
bind_rows(
tibble(country = "33% daily rise", days_since_100 = 0:30) %>%
mutate(cases = 100 * 1.33^days_since_100)
) %>%
ggplot(aes(days_since_100, cases, color = country_region)) +
geom_hline(yintercept = 100) +
geom_vline(xintercept = 0) +
geom_line(size = 0.8) +
geom_point(pch = 21, size = 1) +
scale_y_continuous(
labels = scales::comma_format(accuracy = 1),
breaks = 10^seq(2, 8),
trans = "log10"
) +
theme_minimal() +
scale_color_manual(values = c(
prismatic::clr_darken(paletteer_d("ggsci::category20_d3"), 0.2)[1:length(highlight)],
"gray70"
)) +
theme(
legend.position = "none",
panel.grid.minor = element_blank(),
panel.background = element_rect(fill = "#FFF1E6"),
panel.spacing = margin(3, 15, 3, 15, "mm")
) +
labs(
x = "Number of days since 100th case",
y = "",
title = "Country by country: how coronavirus case trajectories compare",
subtitle = "Cumulative number of cases, by Number of days since 100th case",
caption = "data source from @www.ft.com"
) +
gghighlight::gghighlight(country_region %in% highlight,
label_key = country_region, use_direct_label = TRUE,
label_params = list(segment.color = NA, nudge_x = 1),
use_group_by = FALSE
)