一个简单的例子
tidyverse-dplyr中的across() across() dplyr中的筛选与汇总计算函数相结合使用,实现强大到匪夷所思的功能。
我们先看一个例子
# A tibble: 344 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
6 Adelie Torgersen 39.3 20.6 190 3650
7 Adelie Torgersen 38.9 17.8 181 3625
8 Adelie Torgersen 39.2 19.6 195 4675
9 Adelie Torgersen 34.1 18.1 193 3475
10 Adelie Torgersen 42 20.2 190 4250
# ℹ 334 more rows
# ℹ 2 more variables: sex <fct>, year <int>
如果我们希望统计数据中每一行的缺失值有多少,那么传统方法要怎么写呢?(当然可以使用naniar来简单的计算,但我们更希望在这里用一些tidy传统的方法说明一些问题)
# A tibble: 1 × 8
na_in_species na_in_island na_in_length na_in_depth na_in_flipper na_in_body
<int> <int> <int> <int> <int> <int>
1 0 0 2 2 2 2
# ℹ 2 more variables: na_in_sex <int>, na_in_year <int>
虽然也计算出来了,但好像有点复杂,如果数据框有很多列,那工作量就太大了。有没有简单的方法?试试summarise_all()
# A tibble: 1 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<int> <int> <int> <int> <int> <int>
1 0 0 2 2 2 2
# ℹ 2 more variables: sex <int>, year <int>
没问题。我希望进一步按照企鹅不同的种类,统计各体征数据的均值,用summarise_if()
# A tibble: 3 × 5
species bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <dbl> <dbl> <dbl> <dbl>
1 Adelie 38.8 18.3 190. 3701.
2 Chinstrap 48.8 18.4 196. 3733.
3 Gentoo 47.5 15.0 217. 5076.
除了结果多了一个year列需要处理到外,一切OK。我还希望在结果中加入每类企鹅的数量。因为summarise_if()
# 计算各类企鹅数量,很简单 df2 <- penguins %>% count ( species ) # 直接用count,连group_by()都给它省略。
df2
# A tibble: 3 × 2
species n
<fct> <int>
1 Adelie 152
2 Chinstrap 68
3 Gentoo 124
# A tibble: 3 × 6
species n bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <int> <dbl> <dbl> <dbl> <dbl>
1 Adelie 152 38.8 18.3 190. 3701.
2 Chinstrap 68 48.8 18.4 196. 3733.
3 Gentoo 124 47.5 15.0 217. 5076.
我们将整个步骤回忆下:
summarise_all()
summarise_if() summarise()
left_join()
没问题,思路清晰,代码简洁。但……还有没有更简单的方法?
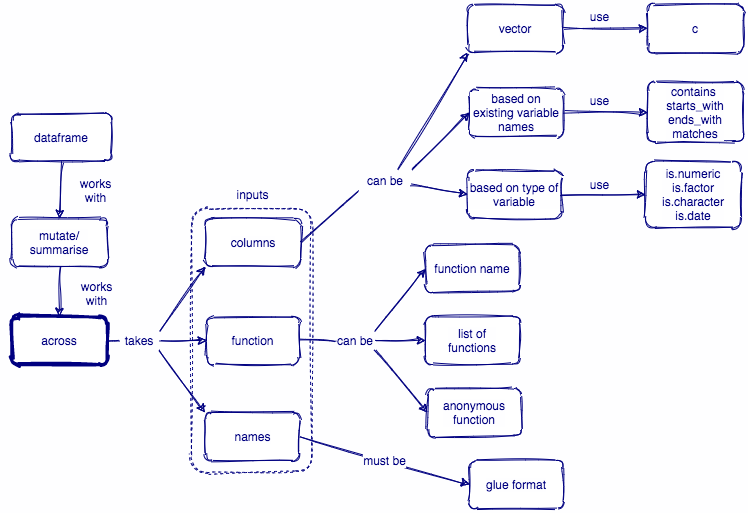
across()函数的基本形式
across() across(.cols, .fns, .nams):
.cols选取列,和select()
.fns,指定对所选列需要执行的函数。可以使默认函数,purrr风格匿名函数,以及一个函数列表(这个列表中也可以同时存在默认函数和匿名函数)。
.names,指定所计算新列的列名,主要针对.fns为一个函数列表时使用。如果.fns是多个函数,就在数据列的列名后面跟上函数名,比如"{.col}_{.fn}";当然,我们也可以简单调整列名和函数之间的顺序或者增加一个标识的字符串,比如弄成"{.fn}_{.col}","{.col}_{.fn}_aa“。
那么现在使用across() summarise_if() summarise()
# A tibble: 1 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<int> <int> <int> <int> <int> <int>
1 0 0 2 2 2 2
# ℹ 2 more variables: sex <int>, year <int>
# A tibble: 3 × 6
species bill_length_mm bill_depth_mm flipper_length_mm body_mass_g n
<fct> <dbl> <dbl> <dbl> <dbl> <int>
1 Adelie 38.8 18.3 190. 3701. 152
2 Chinstrap 48.8 18.4 196. 3733. 68
3 Gentoo 47.5 15.0 217. 5076. 124
好!
across()函数应用
与summarise()连用,对多个列的多个统计量进行统计汇总。
每个类型变量下有多少组
# A tibble: 1 × 3
species island sex
<int> <int> <int>
1 3 3 3
多列多个统计函数
计算每个体征的平均值和标准差。
# A tibble: 3 × 12
species `bill_length_mm-mean` `bill_length_mm-sd` `bill_depth_mm-mean`
<fct> <dbl> <dbl> <dbl>
1 Adelie 38.8 2.66 18.3
2 Chinstrap 48.8 3.34 18.4
3 Gentoo 47.5 3.08 15.0
# ℹ 8 more variables: `bill_depth_mm-sd` <dbl>, `flipper_length_mm-mean` <dbl>,
# `flipper_length_mm-sd` <dbl>, `body_mass_g-mean` <dbl>,
# `body_mass_g-sd` <dbl>, `year-mean` <dbl>, `year-sd` <dbl>, n <int>
不同分组下数据变量的多个分位数
# 传统方法 penguins %>% group_by ( species , island ) %>%
reframe (
prob = c ( 0.25 , 0.75 ) ,
length = quantile ( bill_length_mm , prob , na.rm = TRUE ) ,
depth = quantile ( bill_depth_mm , prob , na.rm = TRUE ) ,
flipper = quantile ( flipper_length_mm , prob , na.rm = TRUE )
)
# A tibble: 10 × 6
species island prob length depth flipper
<fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 Adelie Biscoe 0.25 37.7 17.6 185.
2 Adelie Biscoe 0.75 40.7 19.0 193
3 Adelie Dream 0.25 36.8 17.5 185
4 Adelie Dream 0.75 40.4 18.8 193
5 Adelie Torgersen 0.25 36.7 17.4 187
6 Adelie Torgersen 0.75 41.1 19.2 195
7 Chinstrap Dream 0.25 46.3 17.5 191
8 Chinstrap Dream 0.75 51.1 19.4 201
9 Gentoo Biscoe 0.25 45.3 14.2 212
10 Gentoo Biscoe 0.75 49.6 15.7 221
# A tibble: 10 × 6
species island prob bill_length_mm bill_depth_mm flipper_length_mm
<fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 Adelie Biscoe 0.25 37.7 17.6 185.
2 Adelie Biscoe 0.75 40.7 19.0 193
3 Adelie Dream 0.25 36.8 17.5 185
4 Adelie Dream 0.75 40.4 18.8 193
5 Adelie Torgersen 0.25 36.7 17.4 187
6 Adelie Torgersen 0.75 41.1 19.2 195
7 Chinstrap Dream 0.25 46.3 17.5 191
8 Chinstrap Dream 0.75 51.1 19.4 201
9 Gentoo Biscoe 0.25 45.3 14.2 212
10 Gentoo Biscoe 0.75 49.6 15.7 221
更复杂的统计
将across()
# A tibble: 3 × 6
species n bill_length_mm bill_depth_mm Area body_mass_g
<fct> <int> <dbl> <dbl> <dbl> <dbl>
1 Adelie 152 38.8 18.3 712. 3701.
2 Chinstrap 68 48.8 18.4 900. 3733.
3 Gentoo 124 47.5 15.0 712. 5076.
与mutate()连用-对所有符合条件的列进行函数操作
将数据中小于0的值替换为NA
test <- tibble ( Staff.Confirmed = c ( 0 , 1 , - 999 ) ,
Residents.Confirmed = c ( 12 , - 192 , 0 )
) test
# A tibble: 3 × 2
Staff.Confirmed Residents.Confirmed
<dbl> <dbl>
1 0 12
2 1 -192
3 -999 0
# A tibble: 3 × 4
Staff.Confirmed Residents.Confirmed res_Staff.Confirmed res_Residents.Confir…¹
<dbl> <dbl> <dbl> <dbl>
1 0 12 0 12
2 1 -192 1 NA
3 -999 0 NA 0
# ℹ abbreviated name: ¹res_Residents.Confirmed
对所有符合条件的列操作
# A tibble: 333 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 Adelie Torgersen 3.67 2.93 5.20 8.23
2 Adelie Torgersen 3.68 2.86 5.23 8.24
3 Adelie Torgersen 3.70 2.89 5.27 8.09
4 Adelie Torgersen 3.60 2.96 5.26 8.15
5 Adelie Torgersen 3.67 3.03 5.25 8.20
6 Adelie Torgersen 3.66 2.88 5.20 8.20
7 Adelie Torgersen 3.67 2.98 5.27 8.45
8 Adelie Torgersen 3.72 2.87 5.20 8.07
9 Adelie Torgersen 3.65 3.05 5.25 8.24
10 Adelie Torgersen 3.54 3.05 5.29 8.39
# ℹ 323 more rows
# ℹ 2 more variables: sex <fct>, year <dbl>
与filter()连用-if_all()和if_any()函数
我们通过几个例子来了解下if_any() if_all()
为什么要有这俩函数?因across() filter()
# A tibble: 0 × 8
# ℹ 8 variables: species <fct>, island <fct>, bill_length_mm <dbl>,
# bill_depth_mm <dbl>, flipper_length_mm <int>, body_mass_g <int>, sex <fct>,
# year <int>
筛选有缺失值的行
# A tibble: 11 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen NA NA NA NA
2 Adelie Torgersen 34.1 18.1 193 3475
3 Adelie Torgersen 42 20.2 190 4250
4 Adelie Torgersen 37.8 17.1 186 3300
5 Adelie Torgersen 37.8 17.3 180 3700
6 Adelie Dream 37.5 18.9 179 2975
7 Gentoo Biscoe 44.5 14.3 216 4100
8 Gentoo Biscoe 46.2 14.4 214 4650
9 Gentoo Biscoe 47.3 13.8 216 4725
10 Gentoo Biscoe 44.5 15.7 217 4875
11 Gentoo Biscoe NA NA NA NA
# ℹ 2 more variables: sex <fct>, year <int>
筛选某列大于某值得所有航
# A tibble: 342 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen 36.7 19.3 193 3450
5 Adelie Torgersen 39.3 20.6 190 3650
6 Adelie Torgersen 38.9 17.8 181 3625
7 Adelie Torgersen 39.2 19.6 195 4675
8 Adelie Torgersen 34.1 18.1 193 3475
9 Adelie Torgersen 42 20.2 190 4250
10 Adelie Torgersen 37.8 17.1 186 3300
# ℹ 332 more rows
# ℹ 2 more variables: sex <fct>, year <int>
筛选保留符合条件的行
在指定的列(嘴峰长度和厚度)中检查每行的元素,如果这些元素都大于各自所在列的均值,就保留下来.
# A tibble: 61 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgers… 46 21.5 194 4200
2 Adelie Dream 44.1 19.7 196 4400
3 Adelie Torgers… 45.8 18.9 197 4150
4 Adelie Biscoe 45.6 20.3 191 4600
5 Adelie Torgers… 44.1 18 210 4000
6 Gentoo Biscoe 44.4 17.3 219 5250
7 Gentoo Biscoe 50.8 17.3 228 5600
8 Chinstrap Dream 46.5 17.9 192 3500
9 Chinstrap Dream 50 19.5 196 3900
10 Chinstrap Dream 51.3 19.2 193 3650
# ℹ 51 more rows
# ℹ 2 more variables: sex <fct>, year <int>
在指定的列(嘴峰长度和嘴峰厚度)中检查每行的元素,如果这些元素都大于各自所在列的均值,就“both big”;如果这些元素有一个大于自己所在列的均值,就“one big”,(注意case_when中if_all要在if_any之前 )。
# A tibble: 342 × 9
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen 36.7 19.3 193 3450
5 Adelie Torgersen 39.3 20.6 190 3650
6 Adelie Torgersen 38.9 17.8 181 3625
7 Adelie Torgersen 39.2 19.6 195 4675
8 Adelie Torgersen 34.1 18.1 193 3475
9 Adelie Torgersen 42 20.2 190 4250
10 Adelie Torgersen 37.8 17.1 186 3300
# ℹ 332 more rows
# ℹ 3 more variables: sex <fct>, year <int>, category <chr>
更复杂的例子-across()与迭代
across()
假如我们需要计算一个人每天摄入的水分和食物所占的比例。
library ( tidyverse ) library ( palmerpenguins ) df <- tibble :: tribble ( # tribble 生成rowwise形式的数据框 ~ water , ~ food ,
10.0 , 10.0 ,
12.1 , 10.3 ,
13.5 , 19.1 ,
17.4 , 16.0 ,
25.8 , 15.6 ,
27.4 , 19.8
) df
# A tibble: 6 × 2
water food
<dbl> <dbl>
1 10 10
2 12.1 10.3
3 13.5 19.1
4 17.4 16
5 25.8 15.6
6 27.4 19.8
如果是传统思路,我们应该怎么做?
# A tibble: 6 × 5
rowname water_value food_value water_percent food_percent
<chr> <dbl> <dbl> <dbl> <dbl>
1 1 10 10 50 50
2 2 12.1 10.3 54.0 46.0
3 3 13.5 19.1 41.4 58.6
4 4 17.4 16 52.1 47.9
5 5 25.8 15.6 62.3 37.7
6 6 27.4 19.8 58.1 41.9
使用across()解决以上问题
# A tibble: 6 × 4
water food `porb(%water)` `porb(%food)`
<dbl> <dbl> <dbl> <dbl>
1 10 10 50 50
2 12.1 10.3 54.0 46.0
3 13.5 19.1 41.4 58.6
4 17.4 16 52.1 47.9
5 25.8 15.6 62.3 37.7
6 27.4 19.8 58.1 41.9
迭代
rowwise()+across()组合起来,就是一行一行处理+一列一列处理,换句话说就是一个一个处理。
across()+map_dbl():
有点拗口,我们具体来看。
# A tibble: 3 × 4
# Rowwise:
species `2007` `2008` `2009`
<fct> <int> <int> <int>
1 Adelie 50 50 52
2 Chinstrap 26 18 24
3 Gentoo 34 46 44
# across(.cols = , .fns = purrr::map_dbl()) # 用across()就是一列一列的处理, # 此时的一列是vector or list,又可以进入purrr::map_dbl()再次迭代,对这一列的每个元素,执行.fns # 然后across()到下一列 # 循环模式:第一层,一列一列,第二层在每一列里,一个元素到一个元素 palmerpenguins :: penguins %>% group_by ( species , year ) %>%
summarise ( flipper_length_mm = list ( flipper_length_mm ) ) %>%
ungroup ( ) %>%
pivot_wider (
names_from = year ,
values_from = flipper_length_mm
) %>%
mutate (
across ( where ( is.list ) , function ( x ) map_dbl ( x , length ) )
)
# A tibble: 3 × 4
species `2007` `2008` `2009`
<fct> <dbl> <dbl> <dbl>
1 Adelie 50 50 52
2 Chinstrap 26 18 24
3 Gentoo 34 46 44
案例1
# A tibble: 9 × 4
species year bill_length_mm bill_depth_mm
<fct> <int> <list> <list>
1 Adelie 2007 <dbl [50]> <dbl [50]>
2 Adelie 2008 <dbl [50]> <dbl [50]>
3 Adelie 2009 <dbl [52]> <dbl [52]>
4 Chinstrap 2007 <dbl [26]> <dbl [26]>
5 Chinstrap 2008 <dbl [18]> <dbl [18]>
6 Chinstrap 2009 <dbl [24]> <dbl [24]>
7 Gentoo 2007 <dbl [34]> <dbl [34]>
8 Gentoo 2008 <dbl [46]> <dbl [46]>
9 Gentoo 2009 <dbl [44]> <dbl [44]>
# A tibble: 9 × 6
species year bill_length_mm bill_depth_mm bill_length bill_depth
<fct> <int> <list> <list> <dbl> <dbl>
1 Adelie 2007 <dbl [50]> <dbl [50]> 50 50
2 Adelie 2008 <dbl [50]> <dbl [50]> 50 50
3 Adelie 2009 <dbl [52]> <dbl [52]> 52 52
4 Chinstrap 2007 <dbl [26]> <dbl [26]> 26 26
5 Chinstrap 2008 <dbl [18]> <dbl [18]> 18 18
6 Chinstrap 2009 <dbl [24]> <dbl [24]> 24 24
7 Gentoo 2007 <dbl [34]> <dbl [34]> 34 34
8 Gentoo 2008 <dbl [46]> <dbl [46]> 46 46
9 Gentoo 2009 <dbl [44]> <dbl [44]> 44 44
# A tibble: 9 × 6
species year bill_length_mm bill_depth_mm bill_length bill_depth
<fct> <int> <list> <list> <dbl> <dbl>
1 Adelie 2007 <dbl [50]> <dbl [50]> 50 50
2 Adelie 2008 <dbl [50]> <dbl [50]> 50 50
3 Adelie 2009 <dbl [52]> <dbl [52]> 52 52
4 Chinstrap 2007 <dbl [26]> <dbl [26]> 26 26
5 Chinstrap 2008 <dbl [18]> <dbl [18]> 18 18
6 Chinstrap 2009 <dbl [24]> <dbl [24]> 24 24
7 Gentoo 2007 <dbl [34]> <dbl [34]> 34 34
8 Gentoo 2008 <dbl [46]> <dbl [46]> 46 46
9 Gentoo 2009 <dbl [44]> <dbl [44]> 44 44
# A tibble: 9 × 6
species year bill_length_mm bill_depth_mm bill_length bill_depth
<fct> <int> <list> <list> <dbl> <dbl>
1 Adelie 2007 <dbl [50]> <dbl [50]> 50 50
2 Adelie 2008 <dbl [50]> <dbl [50]> 50 50
3 Adelie 2009 <dbl [52]> <dbl [52]> 52 52
4 Chinstrap 2007 <dbl [26]> <dbl [26]> 26 26
5 Chinstrap 2008 <dbl [18]> <dbl [18]> 18 18
6 Chinstrap 2009 <dbl [24]> <dbl [24]> 24 24
7 Gentoo 2007 <dbl [34]> <dbl [34]> 34 34
8 Gentoo 2008 <dbl [46]> <dbl [46]> 46 46
9 Gentoo 2009 <dbl [44]> <dbl [44]> 44 44
案例2
对于以下数据:
df <- tibble ( id = 1 : 10 ,
sex = c ( "m" , "m" , "m" , "f" , "f" , "f" , "m" , "f" , "f" , "m" ) ,
lds1.x = c ( NA , 1 , 0 , 1 , NA , 0 , 0 , NA , 0 , 1 ) ,
lds1.y = c ( 1 , NA , 1 , 1 , 0 , NA , 0 , 3 , NA , 1 ) ,
lds2.x = c ( 2 , 1 , NA , 0 , 0 , NA , 1 , NA , NA , 1 ) ,
lds2.y = c ( 0 , 2 , 2 , NA , NA , 0 , 0 , 3 , 0 , NA )
) df
# A tibble: 10 × 6
id sex lds1.x lds1.y lds2.x lds2.y
<int> <chr> <dbl> <dbl> <dbl> <dbl>
1 1 m NA 1 2 0
2 2 m 1 NA 1 2
3 3 m 0 1 NA 2
4 4 f 1 1 0 NA
5 5 f NA 0 0 NA
6 6 f 0 NA NA 0
7 7 m 0 0 1 0
8 8 f NA 3 NA 3
9 9 f 0 NA NA 0
10 10 m 1 1 1 NA
用across()
# A tibble: 10 × 8
id sex lds1.x lds1.y lds2.x lds2.y lds1 lds2
<int> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 m NA 1 2 0 1 2
2 2 m 1 NA 1 2 1 1
3 3 m 0 1 NA 2 0 2
4 4 f 1 1 0 NA 1 0
5 5 f NA 0 0 NA 0 0
6 6 f 0 NA NA 0 0 0
7 7 m 0 0 1 0 0 1
8 8 f NA 3 NA 3 3 3
9 9 f 0 NA NA 0 0 0
10 10 m 1 1 1 NA 1 1
思路:
在mutate() across()
# A tibble: 10 × 8
id sex lds1.x lds1.y lds2.x lds2.y lds1 lds2
<int> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 m NA 1 2 0 NA 2
2 2 m 1 NA 1 2 1 1
3 3 m 0 1 NA 2 0 NA
4 4 f 1 1 0 NA 1 0
5 5 f NA 0 0 NA NA 0
6 6 f 0 NA NA 0 0 NA
7 7 m 0 0 1 0 0 1
8 8 f NA 3 NA 3 NA NA
9 9 f 0 NA NA 0 0 NA
10 10 m 1 1 1 NA 1 1
# A tibble: 10 × 8
id sex lds1.x lds1.y lds2.x lds2.y lds1 lds2
<int> <chr> <dbl> <dbl> <dbl> <dbl> <lgl> <lgl>
1 1 m NA 1 2 0 TRUE FALSE
2 2 m 1 NA 1 2 FALSE FALSE
3 3 m 0 1 NA 2 FALSE TRUE
4 4 f 1 1 0 NA FALSE FALSE
5 5 f NA 0 0 NA TRUE FALSE
6 6 f 0 NA NA 0 FALSE TRUE
7 7 m 0 0 1 0 FALSE FALSE
8 8 f NA 3 NA 3 TRUE TRUE
9 9 f 0 NA NA 0 FALSE TRUE
10 10 m 1 1 1 NA FALSE FALSE
# A tibble: 10 × 8
id sex lds1.x lds1.y lds2.x lds2.y lds1 lds2
<int> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 m NA 1 2 0 1 2
2 2 m 1 NA 1 2 1 1
3 3 m 0 1 NA 2 0 2
4 4 f 1 1 0 NA 1 0
5 5 f NA 0 0 NA 0 0
6 6 f 0 NA NA 0 0 0
7 7 m 0 0 1 0 0 1
8 8 f NA 3 NA 3 3 3
9 9 f 0 NA NA 0 0 0
10 10 m 1 1 1 NA 1 1
上述例子中的迭代过程为:.x和.y两个across()
# A tibble: 10 × 8
id sex lds1.x lds1.y lds2.x lds2.y lds1 lds2
<int> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 m NA 1 2 0 1 2
2 2 m 1 NA 1 2 1 1
3 3 m 0 1 NA 2 0 2
4 4 f 1 1 0 NA 1 0
5 5 f NA 0 0 NA 0 0
6 6 f 0 NA NA 0 0 0
7 7 m 0 0 1 0 0 1
8 8 f NA 3 NA 3 3 3
9 9 f 0 NA NA 0 0 0
10 10 m 1 1 1 NA 1 1